DEploid
DEploid is designed for deconvoluting mixed genomes with unknown proportions. Traditional ‘phasing’ programs are limited to diploid organisms. Our method modifies Li and Stephen’s (2003) algorithm with Markov chain Monte Carlo (MCMC) approaches, and builds a generic framework that allows haplotype searches in a multiple infection setting.
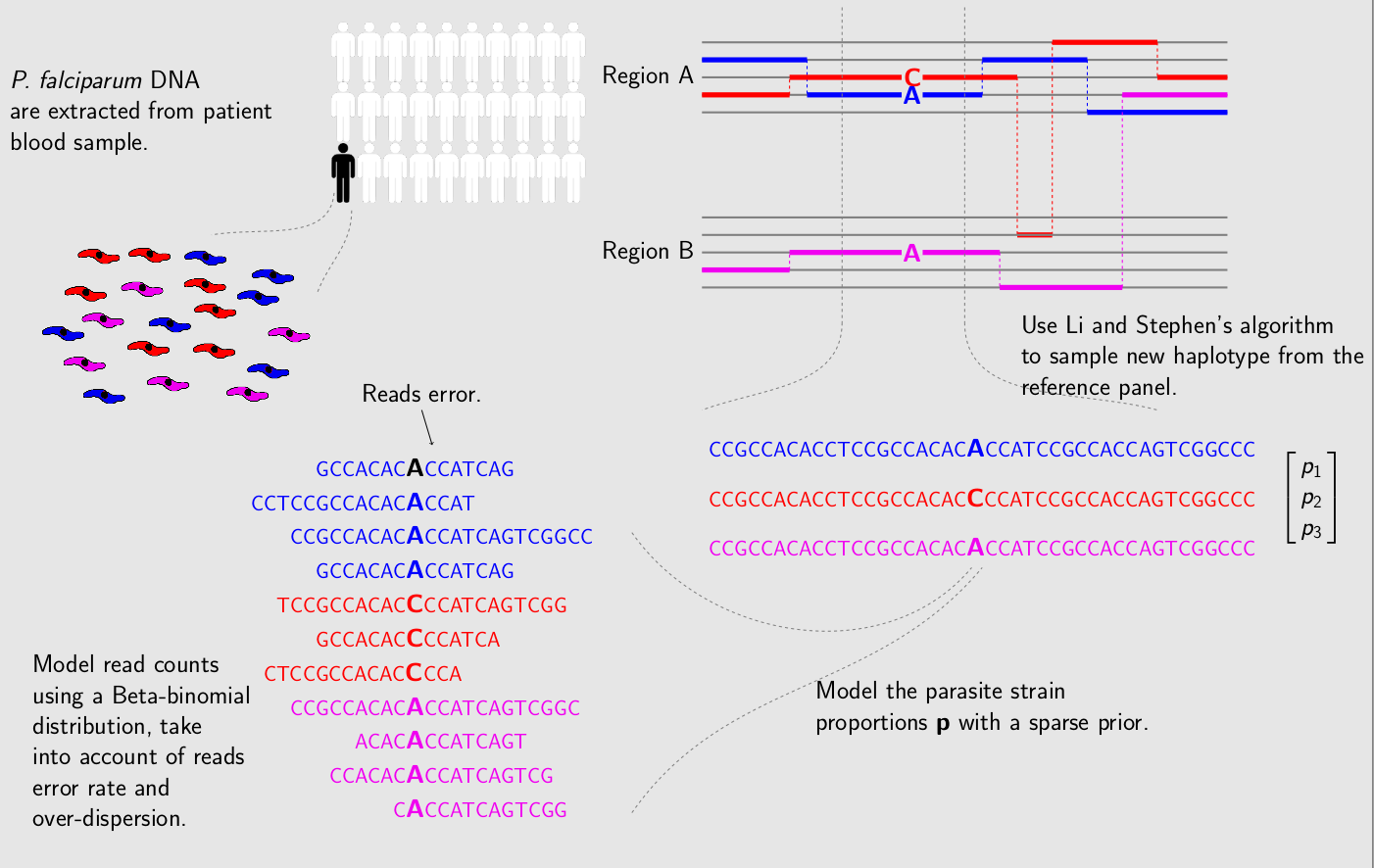
DEploid is primarily developed as part of the Pf3k project, from which this documentation will take examples from for demonstration. The Pf3k project is a global collaboration using the latest sequencing technologies to provide a high-resolution view of natural variation in the malaria parasite Plasmodium falciparum. Parasite DNA are extracted from patient blood sample, which often contains more than one parasite strain, with unknown proportions. DEploid is used for deconvoluting mixed haplotypes, and reporting the mixture proportions from each sample.
Zhu, J. S., J. A. Hendry, J. Almagro-Garcia, R. D. Pearson, R. Amato, A. Miles, D. J. Weiss, T. C. D. Lucas, M. Nguyen, P. W. Gething, D. Kwiatkowski, G. McVean, and for the Pf3k Project. (2018) The origins and relatedness structure of mixed infections vary with local prevalence of P. falciparum malaria. biorxiv, doi: https://doi.org/10.1101/387266.
Zhu, J. S. J. A. Garcia G. McVean. (2018) Deconvolution of multiple infections in Plasmodium falciparum from high throughput sequencing data. Bioinformatics 34(1), 9-15. doi: https://doi.org/10.1093/bioinformatics/btx530.
Evolution of Chinese symbol for ‘parasite’ over 3000 years! #GEM18 @sangerinstitute pic.twitter.com/RK9lwa6ZUP
— CarolineOB (@Caroline_OF_B) June 12, 2018